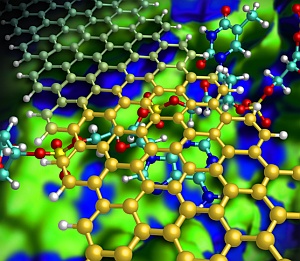
Illustration showing how a graphene nanoribbon (gold) helps to keep a DNA strand (red/blue) flat as it passes through a nanochannel (green/blue).
Nature Nanotechnology – Fast DNA sequencing with a graphene-based nanochannel device
Devices in which a single strand of DNA is threaded through a nanopore could be used to efficiently sequence DNA. However, various issues will have to be resolved to make this approach practical, including controlling the DNA translocation rate, suppressing stochastic nucleobase motions, and resolving the signal overlap between different nucleobases. Here, we demonstrate theoretically the feasibility of DNA sequencing using a fluidic nanochannel functionalized with a graphene nanoribbon. This approach involves deciphering the changes that occur in the conductance of the nanoribbon as a result of its interactions with the nucleobases via π–π stacking. We show that as a DNA strand passes through the nanochannel, the distinct conductance characteristics of the nanoribbon (calculated using a method based on density functional theory coupled to non-equilibrium Green function theory) allow the different nucleobases to be distinguished using a data-mining technique and a two-dimensional transient autocorrelation analysis. This fast and reliable DNA sequencing device should be experimentally feasible in the near future.
DNA base stacking on a graphene nanodevice during its passage through a fluidic nanochannel.
NPG Asia Materials – DNA sequencing: Graphene bridges the gap
Kim and his team designed a hypothetical nanochannel device that radically improves sequencing accuracy by introducing graphene nanoribbons — narrow strips of carbon just one atom thick. In this model (see image), a DNA strand flows down a silicon nitride nanochannel and passes beneath a narrow graphene nanoribbon bridge. Because graphene and nucleotides share similar benzene-like rings, attractive aromatic stacking forces can force the DNA to lie flat in the channel, preventing the random movements of the DNA strand that have previously impeded performance.
The simulations also showed that graphene’s unique electronic properties allow for highly sensitive nucleotide detection. Electrons usually move with zero resistance through the graphene framework, but in the presence of DNA strands, the conductance of the graphene nanoribbon dips sharply following a pattern specific to each nucleotide. The researchers developed a sophisticated algorithm that turned the time-dependent conductance of the device into a precise, automatic read-out of the DNA sequence.
Importantly, the proposed device can be built using existing clean-room protocols, meaning that it may soon be in the hands of researchers. “Identifying a single nucleotide on a graphene nanoribbon would be a significant first step in this direction,” says Kim.
10 pages of supplemental information
If you liked this article, please give it a quick review on ycombinator or StumbleUpon. Thanks

Brian Wang is a Futurist Thought Leader and a popular Science blogger with 1 million readers per month. His blog Nextbigfuture.com is ranked #1 Science News Blog. It covers many disruptive technology and trends including Space, Robotics, Artificial Intelligence, Medicine, Anti-aging Biotechnology, and Nanotechnology.
Known for identifying cutting edge technologies, he is currently a Co-Founder of a startup and fundraiser for high potential early-stage companies. He is the Head of Research for Allocations for deep technology investments and an Angel Investor at Space Angels.
A frequent speaker at corporations, he has been a TEDx speaker, a Singularity University speaker and guest at numerous interviews for radio and podcasts. He is open to public speaking and advising engagements.